Recent advances of -omics technologies have stimulated a large body of biomedical studies focused on the discovery and characterization of molecular mechanisms of various diseases. For example, many studies have been focused on the identification of genes to diagnose or predict cancer. The rapid expansion of complex and large -omics datasets has nourished the development of tailored statistical methods to address the challenges that have arisen in the field. Some examples are detection and correction of biases and artifacts in raw high throughput -omics data, identification of true signal among a large number of variables measured on a much smaller number of subjects, modeling of complex covariance structures, integration of diverse -omics datasets. Research in this area is characterized by multidisciplinary collaborations among researchers from Statistics, Computer Science, Medical Genetics, Molecular Biology, and other related fields.
Recent Highlights
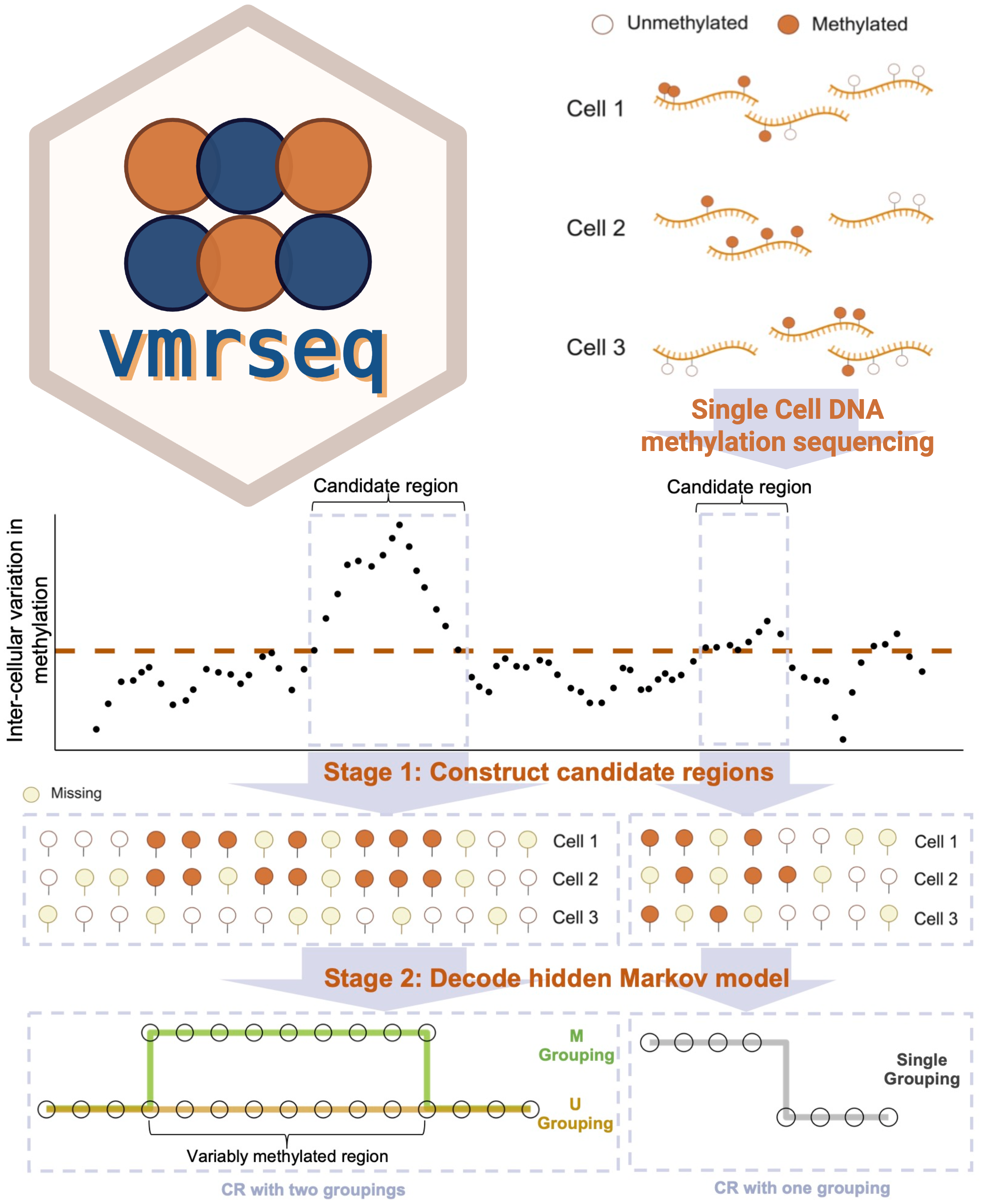

Probabilistic Modeling of Single-Cell DNA Methylation Heterogeneity
K. Korthauer
Recent genomic technologies enable measurement of millions of potential epigenetic modifications to DNA across thousands of individual cells. Interpretation of this high dimensional signal is challenged by its sparsity and complex spatial dependency. To address this challenge, Professor Korthauer and student co-author Shen have recently developed vmrseq, a statistical framework for analyzing single-cell whole genome DNA methylation data that uses hidden Markov modeling and exploits sharing of information across nearby genomic regions and similar cells. The tool enables detection of context-dependent correlations between methylation and gene expression, advancing our understanding of epigenetic gene regulation.
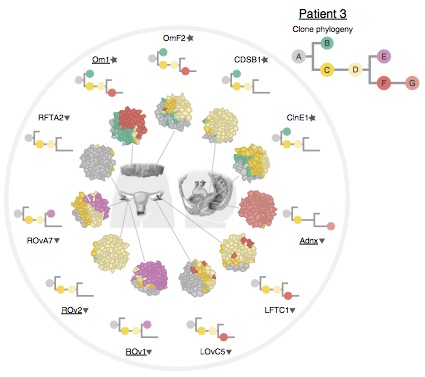
Probabilistic modelling of the evolutionary dynamics and phylogeny of cancer
A. Bouchard-Côté, A. Roth
Proliferating cancer cells, in which DNA repair mechanisms are disrupted, accumulate mutations at a much faster rate than healthy cells do. This leads to the emergence of an evolutionary process inside the tumour. Professor Bouchard-Côté and collaborator Roth are working at the frontier of methodology characterizing the evolutionary dynamics and phylogenies within individual cancer tumours, where multiple sub-populations of cancer cells acquire differentiating sets of mutations.
